Welcome to the ENIGMA-infra guidelines for setting up Nipoppy!
What is Nipoppy?
Nipoppy is a lightweight framework for standardized data organization and processing of neuroimaging-clinical datasets. Its goal is to help users adopt the FAIR principles and improve the reproducibility of studies.
Why?
The figure shows the expected outcomes and corresponding processing steps - most of which can be performed using the Nipoppy framework and helper Python package. We strongly recommend adoption of Nipoppy tools to simplify coordination and ensure reproducibility of this end-to-end process across all sites.
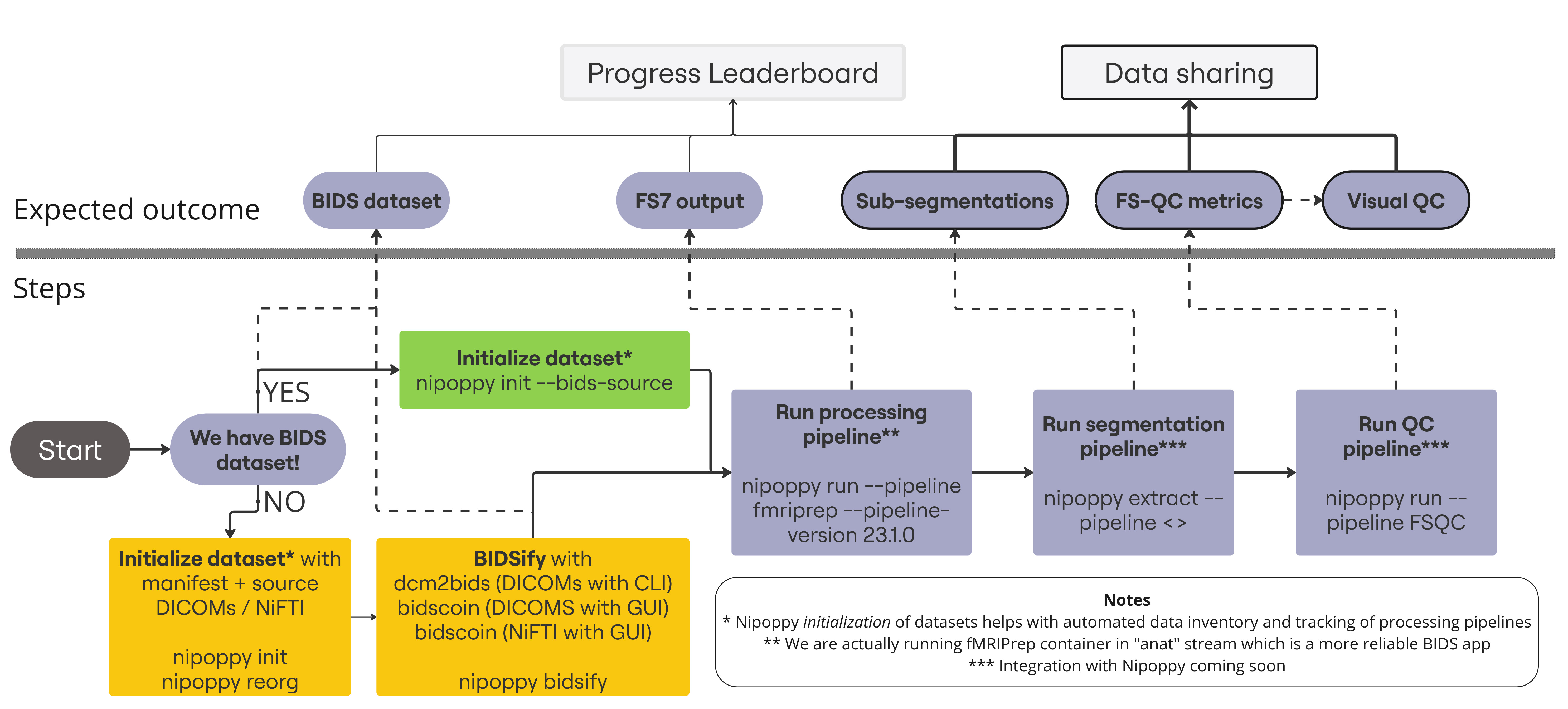
The ongoing collaboration between the ENIGMA working groups and Nipoppy team has streamlined data curation, processing, and analysis workflows, which significantly simplifies tracking of data availability, addition of new pipelines and upgrading of existing pipelines. The ENIGMA and Nipoppy teams are available to support and guide users through the process of implementing the framework, ensuring a smooth transition. To join the Nipoppy support community, we recommend joining the Discord channel. Here you can ask questions and find answers while working with Nipoppy.
In the context of ENIGMA working groups, we will primairly use Nipoppy to help you with BIDSification and to facilitate easy processing with pipelines (such as FreeSurfer processing).
For more information, see the Nipoppy documentation.
Getting started
To install Nipoppy, we refer to the Installation page.
Once Nipoppy is successfully installed, you will need to create a Nipoppy dataset and populate it with your data. There are a few different starting points depending on the current state of your dataset. If you have your data already in BIDS format, click here. If you have DICOM of NIFTI files that are not yet in BIDS, continue below. If you're not sure what BIDS is or if you're wondering why you should convert your data into BIDS at all, you can find more info here.
Starting from source data (either DICOMs or NIfTIs that are not yet in BIDS)
This is the scenario assumed by the Nipoppy Quickstart page. Follow this guide to: 1. Create an empty Nipoppy dataset (i.e. directory tree) 2. Write a manifest file representing your data 3. Modify the global config file with paths to e.g., path to your container directory
Note: if your dataset is cross-sectional (i.e. only has one session), you still need to create a session_id for the manifest. In this case the value would be the same for all participants.
When you reach the end of the Quickstart, it is time to copy and reorganize your raw imaging data to prepare them for BIDS conversion. Once this is done, you can find how to perform the BIDSification within the Nipoppy framework here. We recommend applying a containerized BIDS-conversion pipeline that can be run within Nipoppy. Here you can find how to download containers and here you can find how to run them within Nipoppy.
Starting with BIDSified data
If your dataset is already in BIDS, then the manifest-generation step can be skipped by initializing the Nipoppy dataset with this command, specifying the path to your new dataset and the path to your existing BIDS data:
nipoppy init --dataset <dataset_root> --bids-source <path_to_existing_bids_data>
This command will create a Nipoppy dataset (i.e. directory tree) from preexisting BIDS dataset and automatically generate a manifest file for you!
BIDS datasets without sessions
If the existing BIDS data does not have session-level folders, Nipoppy will create "dummy sessions" (called unnamed) in the manifest. This is because the Nipoppy manifest still requires a non-empty session_id value when imaging data is available for a participant.
If it is feasible to redo the BIDSification to include session folders, we recommend doing so since this is considered good practice. Otherwise, Nipoppy can still be run, but you will need to make some manual changes. For more information, see here
Starting from data already processed with FreeSurfer7
We still encourage you to use Nipoppy to organize your source and/or BIDS data with your processed FS7 output to make use of automated trackers and downstream subsegmentation processing. However, you may need to some help depending on your version of FreeSurfer and naming convention of participant_id. Reach out to us on our Discord channel and we would be happy to help!